What do you get when you mix one part brilliant and one part daft? You get Pylearn2, a cutting edge neural networks library from Montreal that’s rather hard to use. Here we’ll show how to get through the daft part with your mental health relatively intact.
Pylearn2 comes from the Lisa Lab in Montreal, led by Yoshua Bengio. Those are pretty smart guys and they concern themselves with deep learning. Recently they published a paper entitled Pylearn2: a machine learning research library [arxiv]. Here’s a quote:
Pylearn2 is a machine learning research library - its users are researchers. This means (…) it is acceptable to assume that the user has some technical sophistication and knowledge of machine learning.
The word research is possibly the most common word in the paper. There’s a reason for that: the library is certainly not production-ready. OK, it’s not that bad. There are only two difficult things:
- getting your data in
- getting predictions out
What’s attractive about Pylearn2 then? Apparently it’s pretty fast - it’s built on top of Theano, which compiles Python to native code which runs on either CPU or GPU. The downside of this is that compilation takes a while.
The library also features a wealth of deep learning methods, among them the maxout activation function, which is of particular interest to us.
And there’s some documentation. We found the softmax regression tutorial helpful for getting started. Softmax regression is a generalization of logistic regression: instead of two classes, you can have many. We’ll show how to run it and get predictions for the adult dataset.
Getting your data in
To get your data in, you need to write a Python wrapper class for your dataset. Good news: we provide a wrapper for the adult dataset. This wrapper is pretty much ready to be used with other binary classification sets stored as CSV. With slight tweaking of one hot encoding you can use it with multiclass sets.
The wrapper is mainly responsible for loading data. Things like data location and names of training, validation and test sets we prefer to put in the YAML config file.
This is originally made more complicated than it needs to be by get_test_set() method meant to be used in predicting, but we ironed it out a bit by deleting the method. We think it makes more sense to enter a test set path on command line.
Anyway, it seems that most pylearn2 models expect their labels in one-hot encoding, so you need something like this in _load_data():
one_hot = np.zeros(( y.shape[0], self.no_classes ), dtype='float32' )
for i in xrange( y.shape[0] ):
label = y[i]
# a version for two classes
if label == 1:
one_hot[i,1] = 1.
else:
one_hot[i,0] = 1.
y = one_hot
UPDATE: we have written the CSVDataset class for loading data from CSV and got it merged into Pylearn2.
The YAML configuration file
The YAML specifies the job to be done, how to do it and where to save the resulting model. The details are pretty well described in the softmax regression tutorial.
One important thing about YAML is that when you specify a dataset like this:
dataset: &train !obj:adult_dataset.AdultDataset {
The adult_dataset.AdultDataset part refers to a Python file and a Python class in that file. The file should be in your PYTHONPATH, so consider adding a working dir to it:
set PYTHONPATH=%PYTHONPATH%;. or export PYTHONPATH=$PYTHONPATH:.
on Windows and on Unix, respectively.
You can also have your file in a subdirectory of a dir in path:
dataset: &train !obj:first_subdir.second_subdir.adult_dataset.AdultDataset {
...,
with_labels: 0
}
The test set we use has labels; if you’d like to predict on a test set without labels, add with_labels: 0 in the dataset parameters.
Training a model
Time for the easy part:
train.py adult.yaml
During training the library will output a bunch of diagnostics for each set, each epoch:
epochs seen: 5
time trained: 35.890999794
(...)
valid_objective: 0.309834540102
valid_y_col_norms_max: 2.70190905241
valid_y_col_norms_mean: 2.70190905241
valid_y_col_norms_min: 2.70190905241
valid_y_max_max_class: 0.999899923418
valid_y_mean_max_class: 0.866963386567
valid_y_min_max_class: 0.50000807336
valid_y_misclass: 0.148773006135
valid_y_nll: 0.309834540102
valid_y_row_norms_max: 1.10582895147
valid_y_row_norms_mean: 0.259577039679
valid_y_row_norms_min: 0.00150246753658
The most interesting one here would probably be valid_y_misclass - the misclassification rate for the validation set. You can plot the metrics of your choosing:
plot_monitor.py softmax_regression.pkl
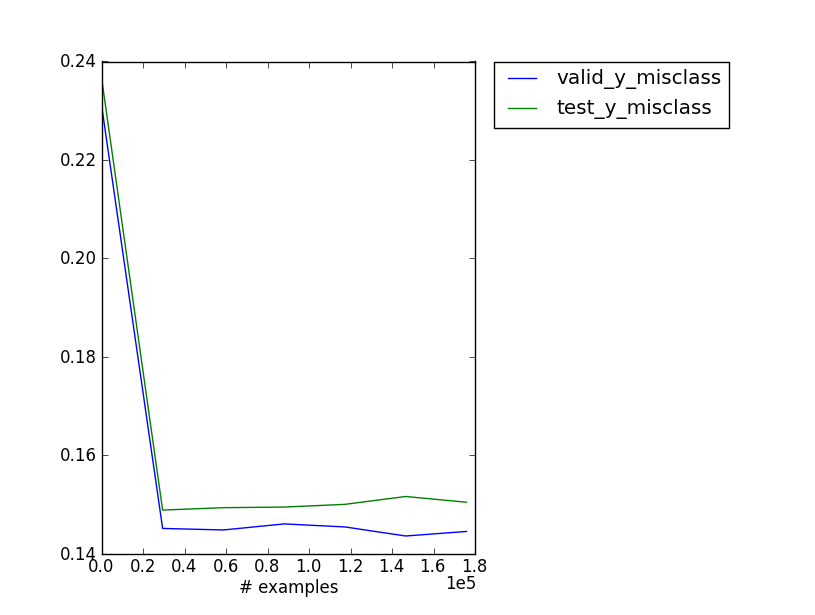
Getting predictions out
This is the hairy part. Pylearn2 is obviously capable of producing predictions - see above. However actually getting those predictions was not paramount in developers’ minds - there’s no single predict script, only a couple of hacks. We modify one of those hacks slightly to suit our needs and that’s it.
predict.py softmax_regression_best.pkl test.csv predictions.txt
First goes a path to a trained model, then a path to a test file, then where you want predictions. Predictions from the model are binary; how to get probabilities is a topic of further study.
The code is available at Github.